The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules
- PMID: 29495629
- PMCID: PMC5867852
- DOI: 10.3390/genes9030131
The Unexplored Diversity of Pleolipoviruses: The Surprising Case of Two Viruses with Identical Major Structural Modules
Abstract
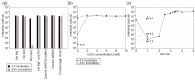
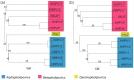
Extremely halophilic Archaea are the only known hosts for pleolipoviruses which are pleomorphic non-lytic viruses resembling cellular membrane vesicles. Recently, pleolipoviruses have been acknowledged by the International Committee on Taxonomy of Viruses (ICTV) as the first virus family that contains related viruses with different DNA genomes. Genomic diversity of pleolipoviruses includes single-stranded and double-stranded DNA molecules and their combinations as linear or circular molecules. To date, only eight viruses belong to the family Pleolipoviridae. In order to obtain more information about the diversity of pleolipoviruses, further isolates are needed. Here we describe the characterization of a new halophilic virus isolate, Haloarcula hispanica pleomorphic virus 4 (HHPV4). All pleolipoviruses and related proviruses contain a conserved core of approximately five genes designating this virus family, but the sequence similarity among different isolates is low. We demonstrate that over half of HHPV4 genome is identical to the genome of pleomorphic virus HHPV3. The genomic regions encoding known virion components are identical between the two viruses, but HHPV4 includes unique genetic elements, e.g., a putative integrase gene. The co-evolution of these two viruses demonstrates the presence of high recombination frequency in halophilic microbiota and can provide new insights considering links between viruses, membrane vesicles, and plasmids.
Keywords: Pleolipoviridae; genomic diversity; haloarchaeal virus; hypersaline environment; pleolipovirus; pleomorphic virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Extremely halophilic pleomorphic archaeal virus HRPV9 extends the diversity of pleolipoviruses with integrases.Res Microbiol. 2018 Nov;169(9):500-504. doi: 10.1016/j.resmic.2018.04.004. Epub 2018 May 14. Res Microbiol. 2018. PMID: 29772256
-
Pleolipoviridae, a newly proposed family comprising archaeal pleomorphic viruses with single-stranded or double-stranded DNA genomes.Arch Virol. 2016 Jan;161(1):249-56. doi: 10.1007/s00705-015-2613-x. Epub 2015 Oct 12. Arch Virol. 2016. PMID: 26459284
-
Vesicle-like virion of Haloarcula hispanica pleomorphic virus 3 preserves high infectivity in saturated salt.Virology. 2016 Dec;499:40-51. doi: 10.1016/j.virol.2016.09.002. Epub 2016 Sep 12. Virology. 2016. PMID: 27632564
-
HCIV-1 and Other Tailless Icosahedral Internal Membrane-Containing Viruses of the Family Sphaerolipoviridae.Viruses. 2017 Feb 18;9(2):32. doi: 10.3390/v9020032. Viruses. 2017. PMID: 28218714 Free PMC article. Review.
-
[The great virus comeback].Biol Aujourdhui. 2013;207(3):153-68. doi: 10.1051/jbio/2013018. Epub 2013 Dec 13. Biol Aujourdhui. 2013. PMID: 24330969 Review. French.
Cited by
-
The Expanding Diversity of Viruses from Extreme Environments.Int J Mol Sci. 2024 Mar 8;25(6):3137. doi: 10.3390/ijms25063137. Int J Mol Sci. 2024. PMID: 38542111 Free PMC article. Review.
-
A Unique m6A-Dependent Restriction Endonuclease from an Archaeal Virus.Microbiol Spectr. 2023 Mar 22;11(2):e0426222. doi: 10.1128/spectrum.04262-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36946751 Free PMC article.
-
A virus-borne DNA damage signaling pathway controls the lysogeny-induction switch in a group of temperate pleolipoviruses.Nucleic Acids Res. 2023 Apr 24;51(7):3270-3287. doi: 10.1093/nar/gkad125. Nucleic Acids Res. 2023. PMID: 36864746 Free PMC article.
-
Growth Phase Dependent Cell Shape of Haloarcula.Microorganisms. 2021 Jan 22;9(2):231. doi: 10.3390/microorganisms9020231. Microorganisms. 2021. PMID: 33499340 Free PMC article.
-
Pleomorphic archaeal viruses: the family Pleolipoviridae is expanding by seven new species.Arch Virol. 2020 Nov;165(11):2723-2731. doi: 10.1007/s00705-020-04689-1. Arch Virol. 2020. PMID: 32583077 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

