EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata
- PMID: 18612431
- PMCID: PMC2440802
- DOI: 10.1371/journal.pone.0002621
EEF2 analysis challenges the monophyly of Archaeplastida and Chromalveolata
Abstract
Background: Classification of eukaryotes provides a fundamental phylogenetic framework for ecological, medical, and industrial research. In recent years eukaryotes have been classified into six major supergroups: Amoebozoa, Archaeplastida, Chromalveolata, Excavata, Opisthokonta, and Rhizaria. According to this supergroup classification, Archaeplastida and Chromalveolata each arose from a single plastid-generating endosymbiotic event involving a cyanobacterium (Archaeplastida) or red alga (Chromalveolata). Although the plastids within members of the Archaeplastida and Chromalveolata share some features, no nucleocytoplasmic synapomorphies supporting these supergroups are currently known.
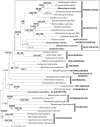
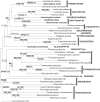
Methodology/principal findings: This study was designed to test the validity of the Archaeplastida and Chromalveolata through the analysis of nucleus-encoded eukaryotic translation elongation factor 2 (EEF2) and cytosolic heat-shock protein of 70 kDa (HSP70) sequences generated from the glaucophyte Cyanophora paradoxa, the cryptophytes Goniomonas truncata and Guillardia theta, the katablepharid Leucocryptos marina, the rhizarian Thaumatomonas sp. and the green alga Mesostigma viride. The HSP70 phylogeny was largely unresolved except for certain well-established groups. In contrast, EEF2 phylogeny recovered many well-established eukaryotic groups and, most interestingly, revealed a well-supported clade composed of cryptophytes, katablepharids, haptophytes, rhodophytes, and Viridiplantae (green algae and land plants). This clade is further supported by the presence of a two amino acid signature within EEF2, which appears to have arisen from amino acid replacement before the common origin of these eukaryotic groups.
Conclusions/significance: Our EEF2 analysis strongly refutes the monophyly of the Archaeplastida and the Chromalveolata, adding to a growing body of evidence that limits the utility of these supergroups. In view of EEF2 phylogeny and other morphological evidence, we discuss the possibility of an alternative eukaryotic supergroup.
Conflict of interest statement
Figures

Similar articles
-
Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates.Mol Biol Evol. 2007 Aug;24(8):1702-13. doi: 10.1093/molbev/msm089. Epub 2007 May 7. Mol Biol Evol. 2007. PMID: 17488740
-
Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes.Mol Biol Evol. 2006 Dec;23(12):2455-66. doi: 10.1093/molbev/msl120. Epub 2006 Sep 18. Mol Biol Evol. 2006. PMID: 16982820
-
Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic "supergroups".Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3859-64. doi: 10.1073/pnas.0807880106. Epub 2009 Feb 23. Proc Natl Acad Sci U S A. 2009. PMID: 19237557 Free PMC article.
-
The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa.Int J Syst Evol Microbiol. 2002 Mar;52(Pt 2):297-354. doi: 10.1099/00207713-52-2-297. Int J Syst Evol Microbiol. 2002. PMID: 11931142 Review.
-
A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the "Plantae," emended.J Plant Res. 2005 Aug;118(4):247-55. doi: 10.1007/s10265-005-0219-1. Epub 2005 Jul 20. J Plant Res. 2005. PMID: 16032387 Review.
Cited by
-
Single cell genomics reveals plastid-lacking Picozoa are close relatives of red algae.Nat Commun. 2021 Nov 17;12(1):6651. doi: 10.1038/s41467-021-26918-0. Nat Commun. 2021. PMID: 34789758 Free PMC article.
-
A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids.Nat Commun. 2021 Mar 25;12(1):1879. doi: 10.1038/s41467-021-22044-z. Nat Commun. 2021. PMID: 33767194 Free PMC article.
-
Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land?Plants (Basel). 2020 Mar 12;9(3):358. doi: 10.3390/plants9030358. Plants (Basel). 2020. PMID: 32178249 Free PMC article.
-
Structure and function of photosystem I in Cyanidioschyzon merolae.Photosynth Res. 2019 Mar;139(1-3):499-508. doi: 10.1007/s11120-018-0501-4. Epub 2018 Mar 26. Photosynth Res. 2019. PMID: 29582227
-
Re-analyses of "Algal" Genes Suggest a Complex Evolutionary History of Oomycetes.Front Plant Sci. 2017 Sep 6;8:1540. doi: 10.3389/fpls.2017.01540. eCollection 2017. Front Plant Sci. 2017. PMID: 28932232 Free PMC article.
References
-
- Bisby FA, Roskov YR, Ruggiero MA, Orrell TM, Paglinawan LE, et al. Species 2000 & ITIS catalogue of life: 2007 annual checklist. Species 2000. Retrieved Jan. 2007. 21, 2008 < www.catalogueoflife.org/annual-checklist/2007/>.
-
- Patterson DJ. The diversity of eukaryotes. Am Nat. 1999;154:S96–S124. - PubMed
-
- Patterson DJ. The lineages of eukaryotes. Tree of life web project. 2000. Retrieved Jan 21, 2008 < www.tolweb.org/notes/note_id49>.
-
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. - PubMed
-
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

