Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription
- PMID: 16282487
- PMCID: PMC1287548
- DOI: 10.1128/JVI.79.23.14876-14886.2005
Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription
Abstract
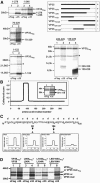
The nucleocapsid protein VP35 of Marburgvirus, a filovirus, acts as the cofactor of the viral polymerase and plays an essential role in transcription and replication of the viral RNA. VP35 forms complexes with the genome encapsidating protein NP and with the RNA-dependent RNA polymerase L. In addition, a trimeric complex had been detected in which VP35 bridges L and the nucleoprotein NP. It has been presumed that the trimeric complex represents the active polymerase bound to the nucleocapsid. Here we present evidence that a predicted coiled-coil domain between amino acids 70 and 120 of VP35 is essential and sufficient to mediate homo-oligomerization of the protein. Substitution of leucine residues 90 and 104 abolished (i) the probability to form coiled coils, (ii) homo-oligomerization, and (iii) the function of VP35 in viral RNA synthesis. Further, it was found that homo-oligomerization-negative mutants of VP35 could not bind to L. Thus, it is presumed that homo-oligomerization-negative mutants of VP35 are unable to recruit the polymerase to the NP/RNA template. In contrast, inability to homo-oligomerize did not abolish the recruitment of VP35 into inclusion bodies, which contain nucleocapsid-like structures formed by NP. Finally, transcriptionally inactive mutants of VP35 containing the functional homo-oligomerization domain displayed a dominant-negative phenotype. Inhibition of VP35 oligomerization might therefore represent a suitable target for antiviral intervention.
Figures

Similar articles
-
Interactions of Marburg virus nucleocapsid proteins.Virology. 1998 Sep 30;249(2):406-17. doi: 10.1006/viro.1998.9328. Virology. 1998. PMID: 9791031
-
Crystal Structure of the Marburg Virus VP35 Oligomerization Domain.J Virol. 2017 Jan 3;91(2):e01085-16. doi: 10.1128/JVI.01085-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847355 Free PMC article.
-
Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus.J Virol. 2017 Jul 27;91(16):e00825-17. doi: 10.1128/JVI.00825-17. Print 2017 Aug 15. J Virol. 2017. PMID: 28566377 Free PMC article.
-
Assembly and transport of filovirus nucleocapsids.PLoS Pathog. 2022 Jul 28;18(7):e1010616. doi: 10.1371/journal.ppat.1010616. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35900983 Free PMC article. Review.
-
Filovirus proteins for antiviral drug discovery: Structure/function bases of the replication cycle.Antiviral Res. 2017 May;141:48-61. doi: 10.1016/j.antiviral.2017.02.004. Epub 2017 Feb 10. Antiviral Res. 2017. PMID: 28192094 Review.
Cited by
-
Ebola and Marburg virus VP35 coiled-coil validated as antiviral target by tripartite split-GFP complementation.iScience. 2022 Oct 13;25(11):105354. doi: 10.1016/j.isci.2022.105354. eCollection 2022 Nov 18. iScience. 2022. PMID: 36325051 Free PMC article.
-
Identification and characterization of coiled-coil motifs across Autographa californica multiple nucleopolyhedrovirus genome.Heliyon. 2022 Sep 10;8(9):e10588. doi: 10.1016/j.heliyon.2022.e10588. eCollection 2022 Sep. Heliyon. 2022. PMID: 36132175 Free PMC article.
-
Functional interactomes of the Ebola virus polymerase identified by proximity proteomics in the context of viral replication.Cell Rep. 2022 Mar 22;38(12):110544. doi: 10.1016/j.celrep.2022.110544. Cell Rep. 2022. PMID: 35320713 Free PMC article.
-
Variation around the dominant viral genome sequence contributes to viral load and outcome in patients with Ebola virus disease.Genome Biol. 2020 Sep 7;21(1):238. doi: 10.1186/s13059-020-02148-3. Genome Biol. 2020. PMID: 32894206 Free PMC article.
-
Structural and Functional Characterization of the Phosphoprotein Central Domain of Spring Viremia of Carp Virus.J Virol. 2020 Jul 16;94(15):e00855-20. doi: 10.1128/JVI.00855-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32434890 Free PMC article.
References
-
- Asenjo, A., and N. Villanueva. 2000. Regulated but not constitutive human respiratory syncytial virus (HRSV) P protein phosphorylation is essential for oligomerization. FEBS Lett. 467:279-284. - PubMed
-
- Becker, S., and E. Mühlberger. 1999. Co- and posttranslational modifications and functions of Marburg virus proteins. Curr. Top. Microbiol. Immunol. 235:23-34. - PubMed
-
- Becker, S., C. Rinne, U. Hofsäss, H.-D. Klenk, and E. Mühlberger. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406-417. - PubMed
-
- Centers for Disease Control and Prevention. 2005. Outbreak of Marburg virus hemorrhagic fever—Angola, October 1, 2004-March 29, 2005. Morb. Mortal. Wkly. Rep. 54:308-309. - PubMed
-
- Choudhary, S. K., A. G. Malur, Y. Huo, B. P. De, and A. K. Banerjee. 2002. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 302:373-382. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

