Improved side-chain modeling for protein-protein docking
- PMID: 15802647
- PMCID: PMC2253276
- DOI: 10.1110/ps.041222905
Improved side-chain modeling for protein-protein docking
Abstract
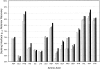
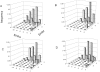
Success in high-resolution protein-protein docking requires accurate modeling of side-chain conformations at the interface. Most current methods either leave side chains fixed in the conformations observed in the unbound protein structures or allow the side chains to sample a set of discrete rotamer conformations. Here we describe a rapid and efficient method for sampling off-rotamer side-chain conformations by torsion space minimization during protein-protein docking starting from discrete rotamer libraries supplemented with side-chain conformations taken from the unbound structures, and show that the new method improves side-chain modeling and increases the energetic discrimination between good and bad models. Analysis of the distribution of side-chain interaction energies within and between the two protein partners shows that the new method leads to more native-like distributions of interaction energies and that the neglect of side-chain entropy produces a small but measurable increase in the number of residues whose interaction energy cannot compensate for the entropic cost of side-chain freezing at the interface. The power of the method is highlighted by a number of predictions of unprecedented accuracy in the recent CAPRI (Critical Assessment of PRedicted Interactions) blind test of protein-protein docking methods.
Figures





Similar articles
-
Progress in protein-protein docking: atomic resolution predictions in the CAPRI experiment using RosettaDock with an improved treatment of side-chain flexibility.Proteins. 2005 Aug 1;60(2):187-94. doi: 10.1002/prot.20556. Proteins. 2005. PMID: 15981249
-
Rapid refinement of protein interfaces incorporating solvation: application to the docking problem.J Mol Biol. 1998 Feb 13;276(1):265-85. doi: 10.1006/jmbi.1997.1519. J Mol Biol. 1998. PMID: 9514726
-
The energetics of off-rotamer protein side-chain conformations.J Mol Biol. 2001 Oct 5;312(5):1161-75. doi: 10.1006/jmbi.2001.4965. J Mol Biol. 2001. PMID: 11580256
-
High-resolution protein-protein docking.Curr Opin Struct Biol. 2006 Apr;16(2):183-93. doi: 10.1016/j.sbi.2006.03.003. Epub 2006 Mar 20. Curr Opin Struct Biol. 2006. PMID: 16546374 Review.
-
Rotamer Dynamics: Analysis of Rotamers in Molecular Dynamics Simulations of Proteins.Biophys J. 2019 Jun 4;116(11):2062-2072. doi: 10.1016/j.bpj.2019.04.017. Epub 2019 Apr 22. Biophys J. 2019. PMID: 31084902 Free PMC article. Review.
Cited by
-
Scientific benchmarks for guiding macromolecular energy function improvement.Methods Enzymol. 2013;523:109-43. doi: 10.1016/B978-0-12-394292-0.00006-0. Methods Enzymol. 2013. PMID: 23422428 Free PMC article.
-
Improved energy bound accuracy enhances the efficiency of continuous protein design.Proteins. 2015 Jun;83(6):1151-64. doi: 10.1002/prot.24808. Epub 2015 May 8. Proteins. 2015. PMID: 25846627 Free PMC article.
-
Structural communication between the GTPase Sec4p and its activator Sec2p: Determinants of GEF activity and early deformations to nucleotide release.Comput Struct Biotechnol J. 2022 Sep 13;20:5162-5180. doi: 10.1016/j.csbj.2022.09.016. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36187918 Free PMC article.
-
Beyond rotamers: a generative, probabilistic model of side chains in proteins.BMC Bioinformatics. 2010 Jun 5;11:306. doi: 10.1186/1471-2105-11-306. BMC Bioinformatics. 2010. PMID: 20525384 Free PMC article.
-
Crystal structure of the HSV-1 Fc receptor bound to Fc reveals a mechanism for antibody bipolar bridging.PLoS Biol. 2006 Jun;4(6):e148. doi: 10.1371/journal.pbio.0040148. Epub 2006 May 2. PLoS Biol. 2006. PMID: 16646632 Free PMC article.
References
-
- Camacho, C.J. and Vajda, S. 2002. Protein–protein association kinetics and protein docking. Curr. Opin. Struct. Biol. 12 36–40. - PubMed
-
- Chen, R. and Weng, Z. 2002. Docking unbound proteins using shape complementarity, desolvation, and electrostatics. Proteins 47 281–294. - PubMed
-
- Chen, R., Mintseris, J., Janin, J., and Weng, Z. 2003. A protein–protein docking benchmark. Proteins 52 88–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

