ClassifieR 2.0: expanding interactive gene expression-based stratification to prostate and high-grade serous ovarian cancer
- PMID: 39574035
- PMCID: PMC11580654
- DOI: 10.1186/s12859-024-05981-6
ClassifieR 2.0: expanding interactive gene expression-based stratification to prostate and high-grade serous ovarian cancer
Abstract
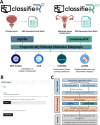
Background: Advances in transcriptional profiling methods have enabled the discovery of molecular subtypes within and across traditional tissue-based cancer classifications. Such molecular subgroups hold potential for improving patient outcomes by guiding treatment decisions and revealing physiological distinctions and targetable pathways. Computational methods for stratifying transcriptomic data into molecular subgroups are increasingly abundant. However, assigning samples to these subtypes and other transcriptionally inferred predictions is time-consuming and requires significant bioinformatics expertise. To address this need, we recently reported "ClassifieR," a flexible, interactive cloud application for the functional annotation of colorectal and breast cancer transcriptomes. Here, we report "ClassifieR 2.0" which introduces additional modules for the molecular subtyping of prostate and high-grade serous ovarian cancer (HGSOC).
Results: ClassifieR 2.0 introduces ClassifieRp and ClassifieRov, two specialised modules specifically designed to address the challenges of prostate and HGSOC molecular classification. ClassifieRp includes sigInfer, a method we developed to infer commercial prognostic prostate gene expression signatures from publicly available gene-lists or indeed any user-uploaded gene-list. ClassifieRov utilizes consensus molecular subtyping methods for HGSOC, including tools like consensusOV, for accurate ovarian cancer stratification. Both modules include functionalities present in the original ClassifieR framework for estimating cellular composition, predicting transcription factor (TF) activity and single sample gene set enrichment analysis (ssGSEA).
Conclusions: ClassifieR 2.0 combines molecular subtyping of prostate cancer and HGSOC with commonly used sample annotation tools in a single, user-friendly platform, allowing scientists without bioinformatics training to explore prostate and HGSOC transcriptional data without the need for extensive bioinformatics knowledge or manual data handling to operate various packages. Our sigInfer method within ClassifieRp enables the inference of commercially available gene signatures for prostate cancer, while ClassifieRov incorporates consensus molecular subtyping for HGSOC. Overall, ClassifieR 2.0 aims to make molecular subtyping more accessible to the wider research community. This is crucial for increased understanding of the molecular heterogeneity of these cancers and developing personalised treatment strategies.
Keywords: High-grade serous ovarian cancer; Molecular classification; Prostate cancer; Shiny application; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: SSM and GPQ are share-holders of generatR Ltd trading as BlokBio, a cloud genomics data analysis company. All other authors have no competing interests.
Figures
Similar articles
-
classifieR a flexible interactive cloud-application for functional annotation of cancer transcriptomes.BMC Bioinformatics. 2022 Mar 31;23(1):114. doi: 10.1186/s12859-022-04641-x. BMC Bioinformatics. 2022. PMID: 35361119 Free PMC article.
-
Platform-Independent Classification System to Predict Molecular Subtypes of High-Grade Serous Ovarian Carcinoma.JCO Clin Cancer Inform. 2019 Apr;3:1-9. doi: 10.1200/CCI.18.00096. JCO Clin Cancer Inform. 2019. PMID: 31002564 Free PMC article.
-
Distinct transcriptional programs stratify ovarian cancer cell lines into the five major histological subtypes.Genome Med. 2021 Sep 1;13(1):140. doi: 10.1186/s13073-021-00952-5. Genome Med. 2021. PMID: 34470661 Free PMC article.
-
Ovarian cancer and the evolution of subtype classifications using transcriptional profiling†.Biol Reprod. 2019 Sep 1;101(3):645-658. doi: 10.1093/biolre/ioz099. Biol Reprod. 2019. PMID: 31187121 Review.
-
Potential Transcriptomic Biomarkers for Predicting Platinum-based Chemotherapy Resistance in Patients With High-grade Serous Ovarian Cancer.Anticancer Res. 2024 Nov;44(11):4691-4707. doi: 10.21873/anticanres.17296. Anticancer Res. 2024. PMID: 39477310 Review.
References
-
- Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. The Breast. 2015;1(24):S26-35. - PubMed
-
- Bijlsma MF, Sadanandam A, Tan P, Vermeulen L. Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2017;14(6):333–42. - PubMed
-
- Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2019;16(4):207–20. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

