A simple and cost-effective transformation system for Porphyromonas gingivalis via natural competence
- PMID: 39498132
- PMCID: PMC11532111
- DOI: 10.3389/fmicb.2024.1476171
A simple and cost-effective transformation system for Porphyromonas gingivalis via natural competence
Abstract
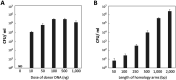
Porphyromonas gingivalis is a major oral bacterial pathogen responsible for severe periodontal diseases. Numerous studies have used genetic approaches to elucidate the molecular mechanisms underlying its pathogenicity. Typically, electroporation and conjugation are utilized for mutagenesis of P. gingivalis; however, these techniques require specialized equipment such as high-voltage electroporators, conjugative plasmids and donor strains. In this study, we present a simple, cost-effective transformation method for P. gingivalis without any special equipment by exploiting its natural DNA competence. P. gingivalis ATCC 33277 was grown to the early-exponential phase and mixed with a donor DNA cassette. This mixture was then spotted onto a BHI-HM blood-agar plate and incubated for one day to promote colony biofilm formation. The resulting colony biofilm was suspended in a liquid medium and spread onto antibiotic-containing agar plates. Transformants appeared within 4 to 5 days, achieving a maximum efficiency of 7.7 × 106 CFU/μg. Although we optimized the transformation conditions using a representative strain ATCC 33277, but the method was also effective for other P. gingivalis strains, W83 and TDC60. Additionally, we discovered that deletion of PGN_0421 or PGN_0519, encoding putative ComEA and ComEC, abolished competency, indicating that these gene products are essential for the natural competence.
Keywords: Porphyromonas gingivalis; genetic engineering; horizontal gene transfer; natural competence; transformation.
Copyright © 2024 Abe, Yahara, Nakao, Yamaguchi and Akeda.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Natural competence is a major mechanism for horizontal DNA transfer in the oral pathogen Porphyromonas gingivalis.mBio. 2012 Jan 31;3(1):e00231-11. doi: 10.1128/mBio.00231-11. Print 2012. mBio. 2012. PMID: 22294679 Free PMC article.
-
Erratum: High-Throughput Identification of Resistance to Pseudomonas syringae pv. Tomato in Tomato using Seedling Flood Assay.J Vis Exp. 2023 Oct 18;(200). doi: 10.3791/6576. J Vis Exp. 2023. PMID: 37851522
-
Characterization of the Porphyromonas gingivalis conjugative transposon CTnPg1: determination of the integration site and the genes essential for conjugal transfer.Microbiology (Reading). 2011 Jul;157(Pt 7):2022-2032. doi: 10.1099/mic.0.047803-0. Epub 2011 Apr 28. Microbiology (Reading). 2011. PMID: 21527470
-
Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis.J Bacteriol. 2007 Sep;189(17):6382-8. doi: 10.1128/JB.00460-07. Epub 2007 Jun 15. J Bacteriol. 2007. PMID: 17573478 Free PMC article.
-
Genetic transformation of Porphyromonas gingivalis by electroporation.Oral Microbiol Immunol. 1993 Aug;8(4):208-12. doi: 10.1111/j.1399-302x.1993.tb00561.x. Oral Microbiol Immunol. 1993. PMID: 8247607
References
Grants and funding
LinkOut - more resources
Full Text Sources

