A genomic hotspot of diversifying selection and structural change in the hoary bat (Lasiurus cinereus)
- PMID: 38832043
- PMCID: PMC11146322
- DOI: 10.7717/peerj.17482
A genomic hotspot of diversifying selection and structural change in the hoary bat (Lasiurus cinereus)
Abstract
Background: Previous work found that numerous genes positively selected within the hoary bat (Lasiurus cinereus) lineage are physically clustered in regions of conserved synteny. Here I further validate and expand on those finding utilizing an updated L. cinereus genome assembly and additional bat species as well as other tetrapod outgroups.
Methods: A chromosome-level assembly was generated by chromatin-contact mapping and made available by DNAZoo (www.dnazoo.org). The genomic organization of orthologous genes was extracted from annotation data for multiple additional bat species as well as other tetrapod clades for which chromosome-level assemblies were available from the National Center for Biotechnology Information (NCBI). Tests of branch-specific positive selection were performed for L. cinereus using PAML as well as with the HyPhy package for comparison.
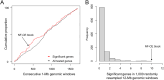
Results: Twelve genes exhibiting significant diversifying selection in the L. cinereus lineage were clustered within a 12-Mb genomic window; one of these (Trpc4) also exhibited diversifying selection in bats generally. Ten of the 12 genes are landmarks of two distinct blocks of ancient synteny that are not linked in other tetrapod clades. Bats are further distinguished by frequent structural rearrangements within these synteny blocks, which are rarely observed in other Tetrapoda. Patterns of gene order and orientation among bat taxa are incompatible with phylogeny as presently understood, implying parallel evolution or subsequent reversals. Inferences of positive selection were found to be robust to alternative phylogenetic topologies as well as a strong shift in background nucleotide composition in some taxa.
Discussion: This study confirms and further localizes a genomic hotspot of protein-coding divergence in the hoary bat, one that also exhibits an increased tempo of structural change in bats compared with other mammals. Most genes in the two synteny blocks have elevated expression in brain tissue in humans and model organisms, and genetic studies implicate the selected genes in cranial and neurological development, among other functions.
Keywords: Adaptation; Comparative genomics; Evolutionary rate analysis; Hoary bat.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Positively selected genes in the hoary bat (Lasiurus cinereus) lineage: prominence of thymus expression, immune and metabolic function, and regions of ancient synteny.PeerJ. 2022 Mar 17;10:e13130. doi: 10.7717/peerj.13130. eCollection 2022. PeerJ. 2022. PMID: 35317076 Free PMC article.
-
Analysis of Genomic Sequence Data Reveals the Origin and Evolutionary Separation of Hawaiian Hoary Bat Populations.Genome Biol Evol. 2020 Sep 1;12(9):1504-1514. doi: 10.1093/gbe/evaa137. Genome Biol Evol. 2020. PMID: 32853363 Free PMC article.
-
Nuclear and mtDNA phylogenetic analyses clarify the evolutionary history of two species of native Hawaiian bats and the taxonomy of Lasiurini (Mammalia: Chiroptera).PLoS One. 2017 Oct 11;12(10):e0186085. doi: 10.1371/journal.pone.0186085. eCollection 2017. PLoS One. 2017. PMID: 29020097 Free PMC article.
-
Geographic origins and population genetics of bats killed at wind-energy facilities.Ecol Appl. 2016 Jul;26(5):1381-1395. doi: 10.1890/15-0541. Ecol Appl. 2016. PMID: 27755755
-
Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX.Nat Protoc. 2024 Jul;19(7):2206-2229. doi: 10.1038/s41596-024-00968-2. Epub 2024 Mar 15. Nat Protoc. 2024. PMID: 38491145 Review.
References
-
- Amador LI, Moyers Arevalo RL, Almeida FC, Catalano SA, Giannini NP. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. Journal of Mammalian Evolution. 2018;25(1):37–70. doi: 10.1007/s10914-016-9363-8. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

