The molecular evolution of spermatogenesis across mammals
- PMID: 36544022
- PMCID: PMC9834047
- DOI: 10.1038/s41586-022-05547-7
The molecular evolution of spermatogenesis across mammals
Abstract
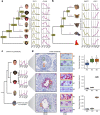
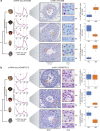
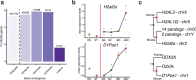
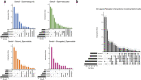
The testis produces gametes through spermatogenesis and evolves rapidly at both the morphological and molecular level in mammals1-6, probably owing to the evolutionary pressure on males to be reproductively successful7. However, the molecular evolution of individual spermatogenic cell types across mammals remains largely uncharacterized. Here we report evolutionary analyses of single-nucleus transcriptome data for testes from 11 species that cover the three main mammalian lineages (eutherians, marsupials and monotremes) and birds (the evolutionary outgroup), and include seven primates. We find that the rapid evolution of the testis was driven by accelerated fixation rates of gene expression changes, amino acid substitutions and new genes in late spermatogenic stages, probably facilitated by reduced pleiotropic constraints, haploid selection and transcriptionally permissive chromatin. We identify temporal expression changes of individual genes across species and conserved expression programs controlling ancestral spermatogenic processes. Genes predominantly expressed in spermatogonia (germ cells fuelling spermatogenesis) and Sertoli (somatic support) cells accumulated on X chromosomes during evolution, presumably owing to male-beneficial selective forces. Further work identified transcriptomal differences between X- and Y-bearing spermatids and uncovered that meiotic sex-chromosome inactivation (MSCI) also occurs in monotremes and hence is common to mammalian sex-chromosome systems. Thus, the mechanism of meiotic silencing of unsynapsed chromatin, which underlies MSCI, is an ancestral mammalian feature. Our study illuminates the molecular evolution of spermatogenesis and associated selective forces, and provides a resource for investigating the biology of the testis across mammals.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Similar articles
-
Evaluating the relationship between spermatogenic silencing of the X chromosome and evolution of the Y chromosome in chimpanzee and human.PLoS One. 2010 Dec 14;5(12):e15598. doi: 10.1371/journal.pone.0015598. PLoS One. 2010. PMID: 21179482 Free PMC article.
-
Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist.J Cell Sci. 2002 Nov 1;115(Pt 21):4097-105. doi: 10.1242/jcs.00111. J Cell Sci. 2002. PMID: 12356914
-
Mechanisms and evolutionary patterns of mammalian and avian dosage compensation.PLoS Biol. 2012;10(5):e1001328. doi: 10.1371/journal.pbio.1001328. Epub 2012 May 15. PLoS Biol. 2012. PMID: 22615540 Free PMC article.
-
The molecular evolution of mammalian spermatogenesis.Cells Dev. 2023 Sep;175:203865. doi: 10.1016/j.cdev.2023.203865. Epub 2023 Jun 17. Cells Dev. 2023. PMID: 37336426 Free PMC article. Review.
-
Mammalian Sex Chromosome Structure, Gene Content, and Function in Male Fertility.Annu Rev Anim Biosci. 2019 Feb 15;7:103-124. doi: 10.1146/annurev-animal-020518-115332. Epub 2018 Nov 9. Annu Rev Anim Biosci. 2019. PMID: 30412673 Review.
Cited by
-
Single-cell transcriptomic and cross-species comparison analyses reveal distinct molecular changes of porcine testes during puberty.Commun Biol. 2024 Nov 9;7(1):1478. doi: 10.1038/s42003-024-07163-9. Commun Biol. 2024. PMID: 39521938 Free PMC article.
-
Evolutionary origins and innovations sculpting the mammalian PRPS enzyme complex.bioRxiv [Preprint]. 2024 Oct 1:2024.10.01.616059. doi: 10.1101/2024.10.01.616059. bioRxiv. 2024. PMID: 39411161 Free PMC article. Preprint.
-
The conserved genetic program of male germ cells uncovers ancient regulators of human spermatogenesis.Elife. 2024 Oct 10;13:RP95774. doi: 10.7554/eLife.95774. Elife. 2024. PMID: 39388236 Free PMC article.
-
Evolution of translational control and the emergence of genes and open reading frames in human and non-human primate hearts.Nat Cardiovasc Res. 2024 Oct;3(10):1217-1235. doi: 10.1038/s44161-024-00544-7. Epub 2024 Sep 24. Nat Cardiovasc Res. 2024. PMID: 39317836 Free PMC article.
-
Adaptive Evolution and Functional Differentiation of Testis-Expressed Genes in Theria.Animals (Basel). 2024 Aug 9;14(16):2316. doi: 10.3390/ani14162316. Animals (Basel). 2024. PMID: 39199849 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

