E2F1-induced lncRNA, EMSLR regulates lncRNA LncPRESS1
- PMID: 35169159
- PMCID: PMC8847401
- DOI: 10.1038/s41598-022-06154-2
E2F1-induced lncRNA, EMSLR regulates lncRNA LncPRESS1
Abstract
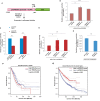
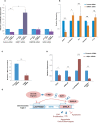
E2F1 induces hundreds of protein-coding genes influencing diverse signaling pathways but much less is known about its non-coding RNA targets. For identifying E2F1-dependent oncogenic long non-coding RNAs (lncRNAs), we carried out genome-wide transcriptome analysis and discovered an lncRNA, EMSLR, which is induced both in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). EMSLR depletion blocks the cells in G1 phase and inhibits the clonogenic ability indicating that it is essential for the tumor-related phenotypes. We discovered that EMSLR represses the promoter activity of another lncRNA, LncPRESS1, which is located 6.9 kb upstream of EMSLR and they display an inverse expression pattern in lung cancer cell lines. Depletion of C-MYC results in downregulation of EMSLR and simultaneous upregulation of EMSLR target LncPRESS1, exemplifying how C-MYC and E2F1 signal transduction pathways control the network of lncRNA genes to modulate cell proliferation and differentiation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Comprehensive Analysis of Aberrantly Expressed Profiles of lncRNAs and miRNAs with Associated ceRNA Network in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma.Pathol Oncol Res. 2020 Jul;26(3):1935-1945. doi: 10.1007/s12253-019-00780-4. Epub 2020 Jan 2. Pathol Oncol Res. 2020. PMID: 31898160
-
LncRNAs are altered in lung squamous cell carcinoma and lung adenocarcinoma.Oncotarget. 2017 Apr 11;8(15):24275-24291. doi: 10.18632/oncotarget.13651. Oncotarget. 2017. PMID: 27903974 Free PMC article.
-
Distinct Patterns of mRNA and lncRNA Expression Differences Between Lung Squamous Cell Carcinoma and Adenocarcinoma.J Comput Biol. 2020 Jul;27(7):1067-1078. doi: 10.1089/cmb.2019.0164. Epub 2019 Nov 22. J Comput Biol. 2020. PMID: 31750732
-
The long non-coding RNA TAZ-AS202 promotes lung cancer progression via regulation of the E2F1 transcription factor and activation of Ephrin signaling.Cell Death Dis. 2023 Nov 18;14(11):752. doi: 10.1038/s41419-023-06277-y. Cell Death Dis. 2023. PMID: 37980331 Free PMC article.
-
Good or not good: Role of miR-18a in cancer biology.Rep Pract Oncol Radiother. 2020 Sep-Oct;25(5):808-819. doi: 10.1016/j.rpor.2020.07.006. Epub 2020 Aug 12. Rep Pract Oncol Radiother. 2020. PMID: 32884453 Free PMC article. Review.
Cited by
-
Current concepts of the crosstalk between lncRNA and E2F1: shedding light on the cancer therapy.Front Pharmacol. 2024 Jul 25;15:1432490. doi: 10.3389/fphar.2024.1432490. eCollection 2024. Front Pharmacol. 2024. PMID: 39119602 Free PMC article. Review.
-
The Role of Long Noncoding RNAs (lncRNAs) in Esophageal Cancer Therapy Resistance and Metastasis.Biomedicines. 2024 Mar 15;12(3):660. doi: 10.3390/biomedicines12030660. Biomedicines. 2024. PMID: 38540273 Free PMC article. Review.
-
Unlocking the Potential of Disulfidptosis-Related LncRNAs in Lung Adenocarcinoma: A Promising Prognostic LncRNA Model for Survival and Immunotherapy Prediction.Cancer Med. 2024 Oct;13(20):e70337. doi: 10.1002/cam4.70337. Cancer Med. 2024. PMID: 39431755 Free PMC article.
-
Prognostic implications of necroptosis-related long noncoding RNA signatures in muscle-invasive bladder cancer.Front Genet. 2022 Dec 2;13:1036098. doi: 10.3389/fgene.2022.1036098. eCollection 2022. Front Genet. 2022. PMID: 36531246 Free PMC article.
-
Disulfidptosis-Related lncRNA for the Establishment of Novel Prognostic Signature and Therapeutic Response Prediction to Endometrial Cancer.Reprod Sci. 2024 Mar;31(3):811-822. doi: 10.1007/s43032-023-01382-x. Epub 2023 Oct 25. Reprod Sci. 2024. PMID: 37880552
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

