A ubiquitous disordered protein interaction module orchestrates transcription elongation
- PMID: 34822292
- PMCID: PMC8943916
- DOI: 10.1126/science.abe2913
A ubiquitous disordered protein interaction module orchestrates transcription elongation
Abstract
During eukaryotic transcription elongation, RNA polymerase II (RNAP2) is regulated by a chorus of factors. Here, we identified a common binary interaction module consisting of TFIIS N-terminal domains (TNDs) and natively unstructured TND-interacting motifs (TIMs). This module was conserved among the elongation machinery and linked complexes including transcription factor TFIIS, Mediator, super elongation complex, elongin, IWS1, SPT6, PP1-PNUTS phosphatase, H3K36me3 readers, and other factors. Using nuclear magnetic resonance, live-cell microscopy, and mass spectrometry, we revealed the structural basis for these interactions and found that TND-TIM sequences were necessary and sufficient to induce strong and specific colocalization in the crowded nuclear environment. Disruption of a single TIM in IWS1 induced robust changes in gene expression and RNAP2 elongation dynamics, which underscores the functional importance of TND-TIM surfaces for transcription elongation.
Conflict of interest statement
Figures




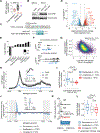
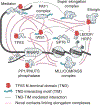
Similar articles
-
The TFIIS N-terminal domain (TND): a transcription assembly module at the interface of order and disorder.Biochem Soc Trans. 2023 Feb 27;51(1):125-135. doi: 10.1042/BST20220342. Biochem Soc Trans. 2023. PMID: 36651856 Free PMC article. Review.
-
The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26.EMBO J. 2010 Dec 1;29(23):3979-91. doi: 10.1038/emboj.2010.272. Epub 2010 Nov 5. EMBO J. 2010. PMID: 21057455 Free PMC article.
-
Structural basis of transcription elongation.Biochim Biophys Acta. 2013 Jan;1829(1):9-19. doi: 10.1016/j.bbagrm.2012.09.002. Epub 2012 Sep 13. Biochim Biophys Acta. 2013. PMID: 22982352 Review.
-
Biophysical Analysis of the N-Terminal Domain from the Human Protein Phosphatase 1 Nuclear Targeting Subunit PNUTS Suggests an Extended Transcription Factor TFIIS-Like Fold.Protein J. 2016 Oct;35(5):340-345. doi: 10.1007/s10930-016-9677-7. Protein J. 2016. PMID: 27591855
-
Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex.Nat Commun. 2017 Jun 6;8:15741. doi: 10.1038/ncomms15741. Nat Commun. 2017. PMID: 28585565 Free PMC article.
Cited by
-
Cardiomyocyte intercellular signalling increases oxidative stress and reprograms the global- and phospho-proteome of cardiac fibroblasts.J Extracell Biol. 2023 Nov 30;2(12):e125. doi: 10.1002/jex2.125. eCollection 2023 Dec. J Extracell Biol. 2023. PMID: 38938901 Free PMC article.
-
Interaction modules that impart specificity to disordered protein.Trends Biochem Sci. 2023 May;48(5):477-490. doi: 10.1016/j.tibs.2023.01.004. Epub 2023 Feb 6. Trends Biochem Sci. 2023. PMID: 36754681 Free PMC article. Review.
-
TFIIS is required for reproductive development and thermal adaptation in barley.Plant Cell Rep. 2024 Oct 10;43(11):260. doi: 10.1007/s00299-024-03345-1. Plant Cell Rep. 2024. PMID: 39390135 Free PMC article.
-
Functional partitioning of transcriptional regulators by patterned charge blocks.Cell. 2023 Jan 19;186(2):327-345.e28. doi: 10.1016/j.cell.2022.12.013. Epub 2023 Jan 4. Cell. 2023. PMID: 36603581 Free PMC article.
-
Suppressor mutations that make the essential transcription factor Spn1/Iws1 dispensable in Saccharomyces cerevisiae.Genetics. 2022 Sep 30;222(2):iyac125. doi: 10.1093/genetics/iyac125. Genetics. 2022. PMID: 35977387 Free PMC article.
References
-
- Miller TE, Liau BB, Wallace LC, Morton AR, Xie Q, Dixit D, Factor DC, Kim LJY, Morrow JJ, Wu Q, Mack SC, Hubert CG, Gillespie SM, Flavahan WA, Hoffmann T, Thummalapalli R, Hemann MT, Paddison PJ, Horbinski CM, Zuber J, Scacheri PC, Bernstein BE, Tesar PJ, Rich JN, Transcription elongation factors represent in vivo cancer dependencies in glioblastoma. Nature (2017), doi:10.1038/nature23000. - DOI - PMC - PubMed
-
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A, AFF4, a Component of the ELL/P-TEFb Elongation Complex and a Shared Subunit of MLL Chimeras, Can Link Transcription Elongation to Leukemia. Mol. Cell (2010), doi:10.1016/j.molcel.2010.01.026. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

