Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer
- PMID: 30154460
- PMCID: PMC6113267
- DOI: 10.1038/s41419-018-0908-z
Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer
Abstract
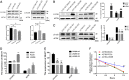
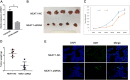
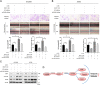
The aberrant expression of long noncoding RNAs (lncRNAs) has been reported frequently in specific cancers, including high-grade serous ovarian cancer (HGSOC). The purpose of the present study was to explore the clinical significance and underlying mechanisms of a significantly dysregulated lncRNA (NEAT1) in HGSOC. Our results showed that elevated NEAT1 expression in human HGSOC specimens correlated with a poor prognosis. Functional experiments demonstrated that knockdown of NEAT1 significantly prohibited ovarian cancer cell proliferation and invasion in vitro and restrained tumor growth in vivo. LIN28B was identified by bioinformatics analysis along with experimental evidence as a direct actor that enhanced NEAT1 stability. A rescue functional assay confirmed that the LIN28B/NEAT1 axis contributed to oncogenic functions in ovarian cancer cells. Moreover, gene expression profile data and dual luciferase reporter assay results demonstrated that NEAT1 functioned as a competing endogenous RNA (ceRNA) for miR-506 to promote cell proliferation and migration. Taken together, our results showed that NEAT1, stabilized by LIN28B, promoted HGSOC progression by sponging miR-506. Thus, NEAT1 can be regarded as a vital diagnostic biomarker for HGSOC and a therapeutic target.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
Long non-coding RNA-NEAT1, a sponge for miR-98-5p, promotes expression of oncogene HMGA2 in prostate cancer.Biosci Rep. 2019 Sep 24;39(9):BSR20190635. doi: 10.1042/BSR20190635. Print 2019 Sep 30. Biosci Rep. 2019. PMID: 31481527 Free PMC article.
-
The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3.J Cell Physiol. 2018 Sep;233(9):6733-6741. doi: 10.1002/jcp.26371. Epub 2018 Apr 16. J Cell Physiol. 2018. PMID: 29219178
-
Long noncoding RNA NEAT1 promotes progression of glioma as a ceRNA by sponging miR-185-5p to stimulate DNMT1/mTOR signaling.J Cell Physiol. 2021 Jan;236(1):121-130. doi: 10.1002/jcp.29644. Epub 2020 Aug 16. J Cell Physiol. 2021. PMID: 32803763
-
An overview of lncRNA NEAT1 contribution in the pathogenesis of female cancers; from diagnosis to therapy resistance.Gene. 2025 Jan 15;933:148975. doi: 10.1016/j.gene.2024.148975. Epub 2024 Sep 29. Gene. 2025. PMID: 39353536 Review.
-
The paradox of autophagy in cancer: NEAT1's role in tumorigenesis and therapeutic resistance.Pathol Res Pract. 2024 Oct;262:155523. doi: 10.1016/j.prp.2024.155523. Epub 2024 Aug 13. Pathol Res Pract. 2024. PMID: 39173466 Review.
Cited by
-
ChrXq27.3 miRNA cluster functions in cancer development.J Exp Clin Cancer Res. 2021 Mar 25;40(1):112. doi: 10.1186/s13046-021-01910-0. J Exp Clin Cancer Res. 2021. PMID: 33766100 Free PMC article. Review.
-
RNA-binding proteins in ovarian cancer: a novel avenue of their roles in diagnosis and treatment.J Transl Med. 2022 Jan 21;20(1):37. doi: 10.1186/s12967-022-03245-6. J Transl Med. 2022. PMID: 35062979 Free PMC article. Review.
-
The role of LIN28B in tumor progression and metastasis in solid tumor entities.Oncol Res. 2023 Apr 10;31(2):101-115. doi: 10.32604/or.2023.028105. eCollection 2023. Oncol Res. 2023. PMID: 37304235 Free PMC article. Review.
-
The pivotal role of ZNF384: driving the malignant behavior of serous ovarian cancer cells via the LIN28B/UBD axis.Cell Biol Toxicol. 2024 Nov 19;40(1):100. doi: 10.1007/s10565-024-09938-6. Cell Biol Toxicol. 2024. PMID: 39562372 Free PMC article.
-
Long non-coding RNA-NEAT1, a sponge for miR-98-5p, promotes expression of oncogene HMGA2 in prostate cancer.Biosci Rep. 2019 Sep 24;39(9):BSR20190635. doi: 10.1042/BSR20190635. Print 2019 Sep 30. Biosci Rep. 2019. PMID: 31481527 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

