Deciphering the role of multiple betaine-carnitine-choline transporters in the Halophile Vibrio parahaemolyticus
- PMID: 25344241
- PMCID: PMC4272712
- DOI: 10.1128/AEM.02402-14
Deciphering the role of multiple betaine-carnitine-choline transporters in the Halophile Vibrio parahaemolyticus
Abstract
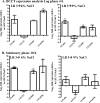
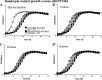
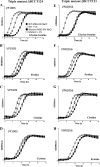
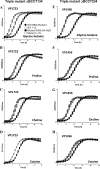
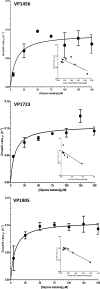
Vibrio parahaemolyticus is a halophile that is the predominant cause of bacterial seafood-related gastroenteritis worldwide. To survive in the marine environment, V. parahaemolyticus must have adaptive strategies to cope with salinity changes. Six putative compatible solute (CS) transport systems were previously predicted from the genome sequence of V. parahaemolyticus RIMD2210633. In this study, we determined the role of the four putative betaine-carnitine-choline transporter (BCCT) homologues VP1456, VP1723, VP1905, and VPA0356 in the NaCl stress response. Expression analysis of the four BCCTs subjected to NaCl upshock showed that VP1456, VP1905, and VPA0356, but not VP1723, were induced. We constructed in-frame single-deletion mutant strains for all four BCCTs, all of which behaved similarly to the wild-type strain, demonstrating a redundancy of the systems. Growth analysis of a quadruple mutant and four BCCT triple mutants demonstrated the requirement for at least one BCCT for efficient CS uptake. We complemented Escherichia coli MHK13, a CS synthesis- and transporter-negative strain, with each BCCT and examined CS uptake by growth analysis and (1)H nuclear magnetic resonance (NMR) spectroscopy analyses. These data demonstrated that VP1456 had the most diverse substrate transport ability, taking up glycine betaine (GB), proline, choline, and ectoine. VP1456 was the sole ectoine transporter. In addition, the data demonstrated that VP1723 can transport GB, proline, and choline, whereas VP1905 and VPA0356 transported only GB. Overall, the data showed that the BCCTs are functional and that there is redundancy among them.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
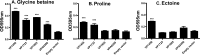

Similar articles
-
Investigations of Dimethylglycine, Glycine Betaine, and Ectoine Uptake by a Betaine-Carnitine-Choline Transporter Family Transporter with Diverse Substrate Specificity in Vibrio Species.J Bacteriol. 2020 Nov 19;202(24):e00314-20. doi: 10.1128/JB.00314-20. Print 2020 Nov 19. J Bacteriol. 2020. PMID: 32817090 Free PMC article.
-
Osmoadaptation among Vibrio species and unique genomic features and physiological responses of Vibrio parahaemolyticus.Appl Environ Microbiol. 2009 May;75(9):2802-10. doi: 10.1128/AEM.01698-08. Epub 2009 Mar 13. Appl Environ Microbiol. 2009. PMID: 19286794 Free PMC article.
-
Marinococcus halophilus DSM 20408T encodes two transporters for compatible solutes belonging to the betaine-carnitine-choline transporter family: identification and characterization of ectoine transporter EctM and glycine betaine transporter BetM.Extremophiles. 2004 Jun;8(3):175-84. doi: 10.1007/s00792-004-0375-6. Epub 2004 Feb 11. Extremophiles. 2004. PMID: 14872322
-
The BCCT family of carriers: from physiology to crystal structure.Mol Microbiol. 2010 Oct;78(1):13-34. doi: 10.1111/j.1365-2958.2010.07332.x. Mol Microbiol. 2010. PMID: 20923416 Review.
-
Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa.Appl Environ Microbiol. 2013 Apr;79(7):2112-20. doi: 10.1128/AEM.03565-12. Epub 2013 Jan 25. Appl Environ Microbiol. 2013. PMID: 23354714 Free PMC article. Review.
Cited by
-
ChoK-ing the Pathogenic Bacteria: Potential of Human Choline Kinase Inhibitors as Antimicrobial Agents.Biomed Res Int. 2020 Jul 9;2020:1823485. doi: 10.1155/2020/1823485. eCollection 2020. Biomed Res Int. 2020. PMID: 32695809 Free PMC article. Review.
-
Three Microbial Musketeers of the Seas: Shewanella baltica, Aliivibrio fischeri and Vibrio harveyi, and Their Adaptation to Different Salinity Probed by a Proteomic Approach.Int J Mol Sci. 2022 Jan 6;23(2):619. doi: 10.3390/ijms23020619. Int J Mol Sci. 2022. PMID: 35054801 Free PMC article.
-
Genomic and transcriptomic analyses reveal distinct biological functions for cold shock proteins (VpaCspA and VpaCspD) in Vibrio parahaemolyticus CHN25 during low-temperature survival.BMC Genomics. 2017 Jun 5;18(1):436. doi: 10.1186/s12864-017-3784-5. BMC Genomics. 2017. PMID: 28583064 Free PMC article.
-
NhaR, LeuO, and H-NS Are Part of an Expanded Regulatory Network for Ectoine Biosynthesis Expression.Appl Environ Microbiol. 2023 Jun 28;89(6):e0047923. doi: 10.1128/aem.00479-23. Epub 2023 Jun 6. Appl Environ Microbiol. 2023. PMID: 37278653 Free PMC article.
-
Investigations of Dimethylglycine, Glycine Betaine, and Ectoine Uptake by a Betaine-Carnitine-Choline Transporter Family Transporter with Diverse Substrate Specificity in Vibrio Species.J Bacteriol. 2020 Nov 19;202(24):e00314-20. doi: 10.1128/JB.00314-20. Print 2020 Nov 19. J Bacteriol. 2020. PMID: 32817090 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

