Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes
- PMID: 23300480
- PMCID: PMC3536677
- DOI: 10.1371/journal.pgen.1003162
Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes
Abstract
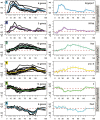
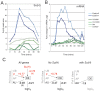
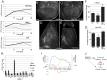
Dynamic activity of signaling pathways, such as Notch, is vital to achieve correct development and homeostasis. However, most studies assess output many hours or days after initiation of signaling, once the outcome has been consolidated. Here we analyze genome-wide changes in transcript levels, binding of the Notch pathway transcription factor, CSL [Suppressor of Hairless, Su(H), in Drosophila], and RNA Polymerase II (Pol II) immediately following a short pulse of Notch stimulation. A total of 154 genes showed significant differential expression (DE) over time, and their expression profiles stratified into 14 clusters based on the timing, magnitude, and direction of DE. E(spl) genes were the most rapidly upregulated, with Su(H), Pol II, and transcript levels increasing within 5-10 minutes. Other genes had a more delayed response, the timing of which was largely unaffected by more prolonged Notch activation. Neither Su(H) binding nor poised Pol II could fully explain the differences between profiles. Instead, our data indicate that regulatory interactions, driven by the early-responding E(spl)bHLH genes, are required. Proposed cross-regulatory relationships were validated in vivo and in cell culture, supporting the view that feed-forward repression by E(spl)bHLH/Hes shapes the response of late-responding genes. Based on these data, we propose a model in which Hes genes are responsible for co-ordinating the Notch response of a wide spectrum of other targets, explaining the critical functions these key regulators play in many developmental and disease contexts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity.Genes Dev. 1995 Nov 1;9(21):2609-22. doi: 10.1101/gad.9.21.2609. Genes Dev. 1995. PMID: 7590239
-
Deadpan contributes to the robustness of the notch response.PLoS One. 2013 Sep 24;8(9):e75632. doi: 10.1371/journal.pone.0075632. eCollection 2013. PLoS One. 2013. PMID: 24086596 Free PMC article.
-
Regulation of expression of Vg and establishment of the dorsoventral compartment boundary in the wing imaginal disc by Suppressor of Hairless.Dev Biol. 2006 Jan 1;289(1):77-90. doi: 10.1016/j.ydbio.2005.10.008. Epub 2005 Nov 22. Dev Biol. 2006. PMID: 16307735
-
E(spl): genetic, developmental, and evolutionary aspects of a group of invertebrate Hes proteins with close ties to Notch signaling.Curr Top Dev Biol. 2014;110:217-62. doi: 10.1016/B978-0-12-405943-6.00006-3. Curr Top Dev Biol. 2014. PMID: 25248478 Review.
-
HES and HERP families: multiple effectors of the Notch signaling pathway.J Cell Physiol. 2003 Mar;194(3):237-55. doi: 10.1002/jcp.10208. J Cell Physiol. 2003. PMID: 12548545 Review.
Cited by
-
Image-Based Single-Molecule Analysis of Notch-Dependent Transcription in Its Natural Context.Methods Mol Biol. 2022;2472:131-149. doi: 10.1007/978-1-0716-2201-8_11. Methods Mol Biol. 2022. PMID: 35674897
-
The MEK/ERK pathway promotes NOTCH signalling in pancreatic cancer cells.PLoS One. 2013 Dec 31;8(12):e85502. doi: 10.1371/journal.pone.0085502. eCollection 2013. PLoS One. 2013. PMID: 24392017 Free PMC article.
-
Inducible defenses stay up late: temporal patterns of immune gene expression in Tenebrio molitor.G3 (Bethesda). 2013 Dec 6;4(6):947-55. doi: 10.1534/g3.113.008516. G3 (Bethesda). 2013. PMID: 24318927 Free PMC article.
-
The Groucho co-repressor is primarily recruited to local target sites in active chromatin to attenuate transcription.PLoS Genet. 2014 Aug 28;10(8):e1004595. doi: 10.1371/journal.pgen.1004595. eCollection 2014 Aug. PLoS Genet. 2014. PMID: 25165826 Free PMC article.
-
Mastermind-Like 1 Is Ubiquitinated: Functional Consequences for Notch Signaling.PLoS One. 2015 Jul 30;10(7):e0134013. doi: 10.1371/journal.pone.0134013. eCollection 2015. PLoS One. 2015. PMID: 26225565 Free PMC article.
References
-
- Akimov V, Rigbolt KT, Nielsen MM, Blagoev B (2011) Characterization of ubiquitination dependent dynamics in growth factor receptor signaling by quantitative proteomics. Molecular BioSystems 7: 3223–3233. - PubMed
-
- Kawahashi K, Hayashi S (2010) Dynamic intracellular distribution of Notch during activation and asymmetric cell division revealed by functional fluorescent fusion proteins. Genes to Cells : Devoted to Molecular & Cellular Mechanisms 15: 749–759. - PubMed
-
- Andersson ER, Sandberg R, Lendahl U (2011) Notch signaling: simplicity in design, versatility in function. Development 138: 3593–3612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

