The canonical Notch signaling pathway: unfolding the activation mechanism
- PMID: 19379690
- PMCID: PMC2827930
- DOI: 10.1016/j.cell.2009.03.045
The canonical Notch signaling pathway: unfolding the activation mechanism
Abstract
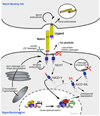
Notch signaling regulates many aspects of metazoan development and tissue renewal. Accordingly, the misregulation or loss of Notch signaling underlies a wide range of human disorders, from developmental syndromes to adult-onset diseases and cancer. Notch signaling is remarkably robust in most tissues even though each Notch molecule is irreversibly activated by proteolysis and signals only once without amplification by secondary messenger cascades. In this Review, we highlight recent studies in Notch signaling that reveal new molecular details about the regulation of ligand-mediated receptor activation, receptor proteolysis, and target selection.
Figures



Similar articles
-
The multiple facets of ubiquitination in the regulation of notch signaling pathway.Traffic. 2011 Feb;12(2):149-61. doi: 10.1111/j.1600-0854.2010.01126.x. Epub 2010 Oct 29. Traffic. 2011. PMID: 21029288 Review.
-
Canonical and non-canonical Notch ligands.Curr Top Dev Biol. 2010;92:73-129. doi: 10.1016/S0070-2153(10)92003-6. Curr Top Dev Biol. 2010. PMID: 20816393 Free PMC article. Review.
-
The molecular logic of Notch signaling--a structural and biochemical perspective.J Cell Sci. 2008 Oct 1;121(Pt 19):3109-19. doi: 10.1242/jcs.035683. J Cell Sci. 2008. PMID: 18799787 Free PMC article. Review.
-
Bare rudiments of notch signaling: how receptor levels are regulated.Trends Biochem Sci. 2007 Oct;32(10):477-85. doi: 10.1016/j.tibs.2007.09.002. Trends Biochem Sci. 2007. PMID: 17920278
-
The Molecular Mechanism of Notch Activation.Adv Exp Med Biol. 2018;1066:47-58. doi: 10.1007/978-3-319-89512-3_3. Adv Exp Med Biol. 2018. PMID: 30030821 Review.
Cited by
-
Transcriptional dynamics elicited by a short pulse of notch activation involves feed-forward regulation by E(spl)/Hes genes.PLoS Genet. 2013;9(1):e1003162. doi: 10.1371/journal.pgen.1003162. Epub 2013 Jan 3. PLoS Genet. 2013. PMID: 23300480 Free PMC article.
-
Human NOTCH2 Is Resistant to Ligand-independent Activation by Metalloprotease Adam17.J Biol Chem. 2015 Jun 5;290(23):14705-16. doi: 10.1074/jbc.M115.643676. Epub 2015 Apr 27. J Biol Chem. 2015. PMID: 25918160 Free PMC article.
-
Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner.Mol Cancer. 2015 Feb 3;14(1):28. doi: 10.1186/s12943-015-0295-3. Mol Cancer. 2015. PMID: 25645291 Free PMC article.
-
Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch.J Biol Chem. 2012 Jan 20;287(4):2739-48. doi: 10.1074/jbc.M111.302406. Epub 2011 Nov 23. J Biol Chem. 2012. PMID: 22117070 Free PMC article.
-
Major pathways involved in macrophage polarization in cancer.Front Immunol. 2022 Oct 17;13:1026954. doi: 10.3389/fimmu.2022.1026954. eCollection 2022. Front Immunol. 2022. PMID: 36325334 Free PMC article. Review.
References
-
- Alves-Guerra MC, Ronchini C, Capobianco AJ. Mastermind-like 1 Is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 2007;67:8690–8698. - PubMed
-
- Ambros V. Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development. 1999;126:1947–1956. - PubMed
-
- Bardin AJ, Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev Cell. 2006;10:245–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

