Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness
- PMID: 15746353
- PMCID: PMC1065144
- DOI: 10.1128/AEM.71.3.1501-1506.2005
Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness
Abstract
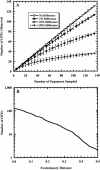
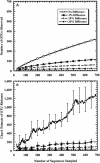
Although copious qualitative information describes the members of the diverse microbial communities on Earth, statistical approaches for quantifying and comparing the numbers and compositions of lineages in communities are lacking. We present a method that addresses the challenge of assigning sequences to operational taxonomic units (OTUs) based on the genetic distances between sequences. We developed a computer program, DOTUR, which assigns sequences to OTUs by using either the furthest, average, or nearest neighbor algorithm for each distance level. DOTUR uses the frequency at which each OTU is observed to construct rarefaction and collector's curves for various measures of richness and diversity. We analyzed 16S rRNA gene libraries derived from Scottish and Amazonian soils and the Sargasso Sea with DOTUR, which assigned sequences to OTUs rapidly and reliably based on the genetic distances between sequences and identified previous inconsistencies and errors in assigning sequences to OTUs. An analysis of the two 16S rRNA gene libraries from soil demonstrated that they do not contain enough sequences to support a claim that they contain different numbers of bacterial lineages with statistical confidence (P > 0.05), nor do they contain enough sequences to provide a robust estimate of species richness when an OTU is defined as containing sequences that are no more than 3% different from each other. In contrast, the richness of OTUs at the 3% level in the Sargasso Sea collection began to plateau after the sampling of 690 sequences. We anticipate that an equivalent extent of sampling for soil would require sampling more than 10,000 sequences, almost 100 times the size of typical sequence collections obtained from soil.
Figures


Similar articles
-
[Diversity analysis of archaeal and fungal communities in adjacent cucumber root soil samples in greenhouse by small-subunit rRNA gene cloning].Sheng Wu Gong Cheng Xue Bao. 2011 Jan;27(1):41-51. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 21553489 Chinese.
-
Toward a census of bacteria in soil.PLoS Comput Biol. 2006 Jul 21;2(7):e92. doi: 10.1371/journal.pcbi.0020092. Epub 2006 Jun 5. PLoS Comput Biol. 2006. PMID: 16848637 Free PMC article.
-
Microbial community of salt crystals processed from Mediterranean seawater based on 16S rRNA analysis.Can J Microbiol. 2010 Jan;56(1):44-51. doi: 10.1139/w09-102. Can J Microbiol. 2010. PMID: 20130693
-
Comparison of bacterial communities in the Solimões and Negro River tributaries of the Amazon River based on small subunit rRNA gene sequences.Genet Mol Res. 2011 Dec 8;10(4):3783-93. doi: 10.4238/2011.December.8.8. Genet Mol Res. 2011. PMID: 22183948
-
Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes.J Microbiol Methods. 2011 Jan;84(1):81-7. doi: 10.1016/j.mimet.2010.10.020. Epub 2010 Oct 31. J Microbiol Methods. 2011. PMID: 21047533
Cited by
-
Dissecting the Simultaneous Extracellular/Intracellular Contributions to Cr(VI) Reduction under Aerobic and Anaerobic Conditions Using the Newly Isolating Cr(VI)-Reducing Bacterium of Pseudomonas sp. HGB10.Microorganisms. 2024 Sep 27;12(10):1958. doi: 10.3390/microorganisms12101958. Microorganisms. 2024. PMID: 39458268 Free PMC article.
-
Occurrence of Uncultured Legionella spp. in Treated Wastewater Effluent and Its Impact on Human Health (SCA.Re.S Project).Pathogens. 2024 Sep 12;13(9):786. doi: 10.3390/pathogens13090786. Pathogens. 2024. PMID: 39338977 Free PMC article.
-
SpeciateIT and vSpeciateDB: novel, fast, and accurate per sequence 16S rRNA gene taxonomic classification of vaginal microbiota.BMC Bioinformatics. 2024 Sep 27;25(1):313. doi: 10.1186/s12859-024-05930-3. BMC Bioinformatics. 2024. PMID: 39333850 Free PMC article.
-
Reproductive Tract Microbial Transitions from Late Gestation to Early Postpartum Using 16S rRNA Metagenetic Profiling in First-Pregnancy Heifers.Int J Mol Sci. 2024 Aug 23;25(17):9164. doi: 10.3390/ijms25179164. Int J Mol Sci. 2024. PMID: 39273112 Free PMC article.
-
Culture-dependent identification of rare marine sediment bacteria from the Gulf of Mexico and Antarctica.bioRxiv [Preprint]. 2024 Jun 11:2024.06.11.598530. doi: 10.1101/2024.06.11.598530. bioRxiv. 2024. PMID: 38915660 Free PMC article. Preprint.
References
-
- Burnham, K. P., and W. S. Overton. 1979. Robust estimation of population size when capture probabilities vary among animals. Ecology 60:927-936.
-
- Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270.
-
- Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

